Example 1: Sparsity-Assisted Signal Smoothing (K = 2)
This example shows the use of SASS for filtering a signal that has discontinuities in its derivative (the signal has cusps). SASS is based on modeling the data, y, as: y = f + g + w where f is a low-pass signal, g has a sparse order-k derivative, and w is white noise.
Ivan Selesnick selesi@poly.edu 2013
Contents
Start
clear printme = @(filename) print('-dpdf', sprintf('figures/Example1_%s', filename)); ramp = @(n) n .* (n >= 0);
Make signals
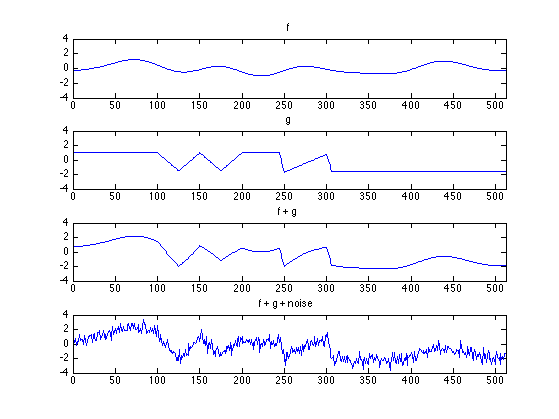
N = 512; n = (0:N-1)'; % M- : discontinuity points M1 = 100; M2 = 125; M3 = 150; M4 = 250; M5 = 250; M6 = 300; M7 = 150; M8 = 175; M9 = 200; % g : piecewise linear signal g = 2 * ramp( n - M1 ) - 4 * ramp( n - M2 ) + 2 * ramp( n - M3 ); g = g + 9 * (ramp(n - M4 + 6) - ramp( n - M4 )); g = g - ramp( n - M5 ) + 9 * ramp( n - M6 ) - 8 * ramp( n - M6 - 6 ); g = g + 2 * ramp( n - M7 ) - 4 * ramp( n - M8 ) + 2*ramp( n - M9 ) ; g = 1 - 0.05 * g; randn('state', 1); rand('state', 1); % set rand/randn states for reproducibility of example f = zeros(size(n)); % f : low-frequency sinusoids for k = 1:6 f = f + 0.5 * randn * cos(2*pi*k/N*n + 2*pi*rand); end sigma = 0.5; noise = sigma * randn(size(n)); % noise : white Gaussian noise s = g + f; % s : noise-free signal data = g + f + noise; % data : noisy signal
Display signals
ax = [0 N -4 4]; figure(1) clf subplot(4, 1, 1) plot(n, f) title('f') axis(ax) subplot(4, 1, 2) plot(n, g); title('g') axis(ax) subplot(4, 1, 3) plot(n, s) title('f + g') axis(ax) subplot(4, 1, 4) plot(n, data) title('f + g + noise') axis(ax) orient tall printme('data')
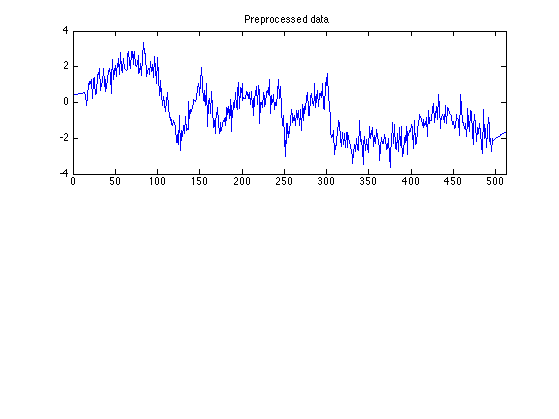
Perform preprocessing
The SASS algorithm may introduce transients at the beginning and end of the signal. To reduce the transients, replace first and last samples of the data by a low-order polynomial approximation.
r = 1; M = 15; y = preproc(r, M, data); % y : preprocessed data figure(1) clf subplot(2, 1, 1) plot(n, y) title('Preprocessed data') xlim([0 N])
Filter matrices
Define filter matrices for SASS.
d = 1; % filter is of order 2d fc = 0.02; % fc : cut-off frequency (0 < fc < 0.5) (cycles/sample) K = 2; % K : order of sparse derivative % Banded filter matrices: [A, B, B1, D, a, b, b1, H1norm HTH1norm] = ABfilt(d, fc, N, K); % [A, B] = ABfilt(d, fc, N, K); H = @(x) [nan(d,1); A\(B*x); nan(d,1)]; % H: high-pass filter L = @(x) x - H(x); % L: low-pass filter txt_filt = sprintf('d = %d, fc = %.3f', d, fc);
Low-pass filter
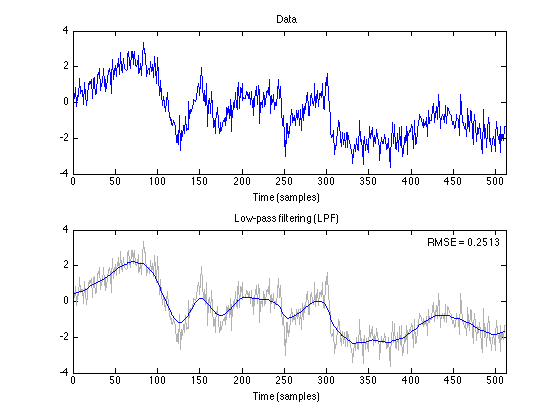
The low-pass filter preserves the low-pass signal, f, with high accuracy, but significantly distorts the piecewise linear signal, g. This guides how to set the cuf-off frequency, fc. fc should be chosen so that the low-pass filter passes the low-pass signal.
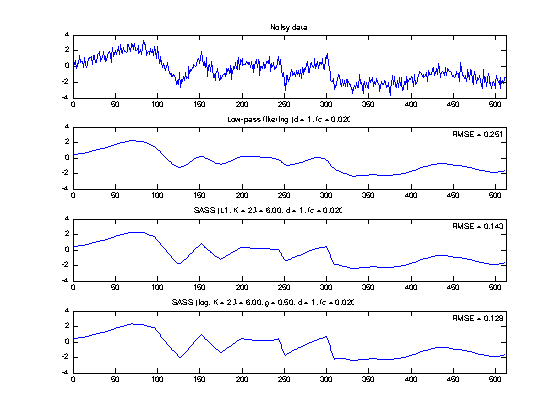
x_lpf = L(y); txt_LPF = sprintf('Low-pass filtering (%s)', txt_filt); err = s - x_lpf; RMSE_LPF = sqrt(mean(err(K+1:end-K).^2)) figure(1) clf GRAY = [1 1 1]*0.7; subplot(2, 1, 1) plot(n, data) title('Data') xlabel('Time (samples)') axis(ax) subplot(2, 1, 2) plot(n, data, 'color', GRAY) line(n, x_lpf); title('Low-pass filtering (LPF)') axis(ax) xlabel('Time (samples)') axnote(sprintf('RMSE = %.4f', RMSE_LPF)) orient portrait printme('LPF')
RMSE_LPF =
0.2513
SASS (L1 norm)
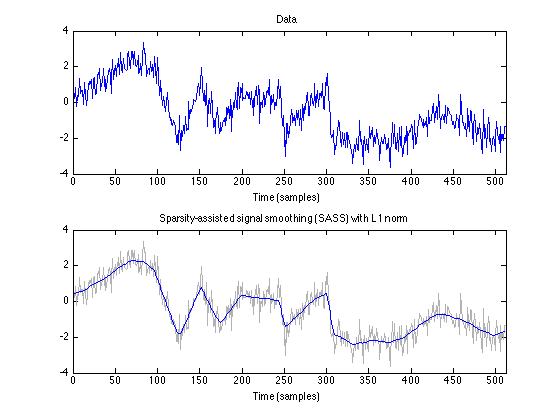
beta = 3; lam = beta * sigma * HTH1norm; % lam : regularization parameter Nit = 100; % Nit : number of iterations [x_L1, cost_L1, u_L1, v_L1] = sass(y, d, fc, K, lam, 'L1', [], Nit); % x : denoised signal % cost : cost function history % u : sparse order-K derivative % v : inv(A)*B1*u txt_L1 = sprintf('SASS (L1, K = %d, \\lambda = %.2f, %s)', K, lam, txt_filt) err = s - x_L1; RMSE_L1 = sqrt(mean(err(K+1:end-K).^2))
0 incorrectly locked zeros detected.
txt_L1 =
SASS (L1, K = 2, \lambda = 6.00, d = 1, fc = 0.020)
RMSE_L1 =
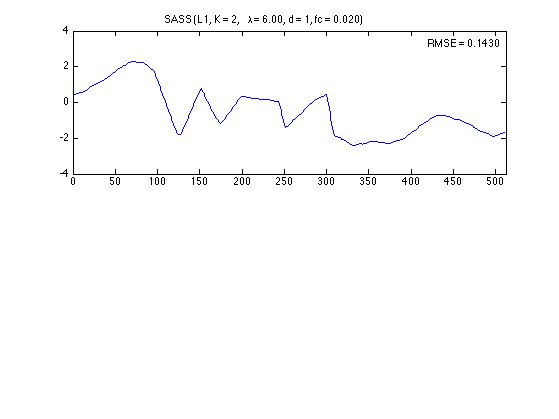
0.1430
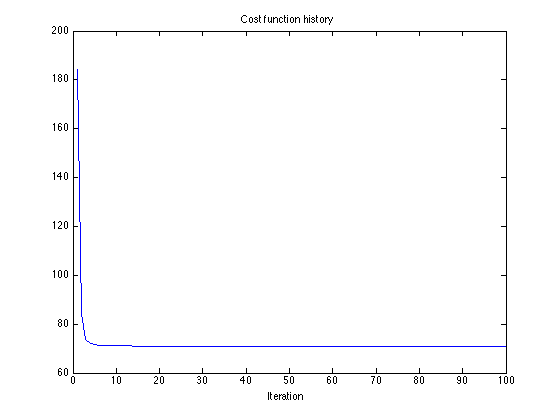
Cost function
figure(2) clf plot(cost_L1) title('Cost function history'); xlabel('Iteration')
Denoised signal
figure(2)
clf
subplot(2, 1, 1)
plot(n, x_L1)
axnote(sprintf('RMSE = %.4f', RMSE_L1))
title(txt_L1)
axis(ax)
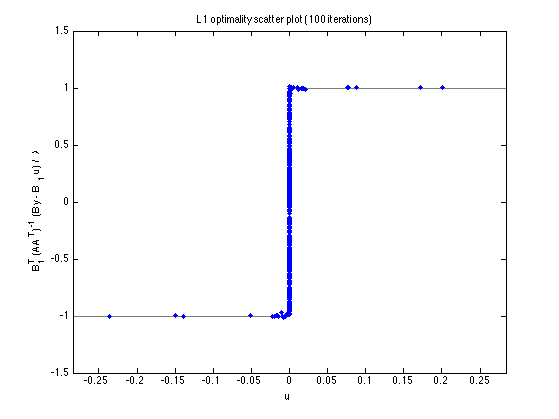
Optimality scatter plot
figure(2) clf plot_scatter(A, B, B1, y, u_L1, lam, 'L1', []); title(sprintf('L1 optimality scatter plot (%d iterations)', Nit)) printme('L1_scatter')
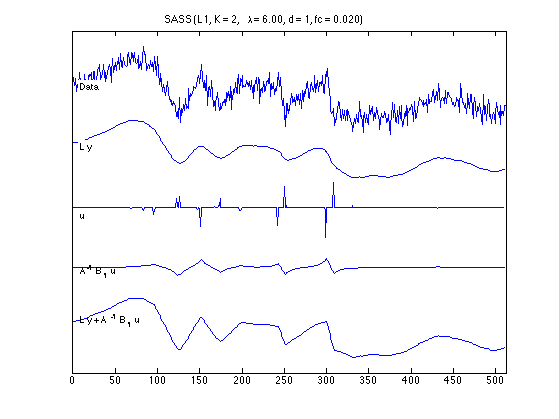
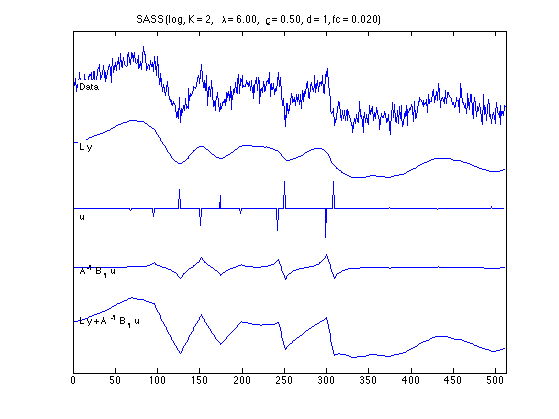
Components
figure(1) clf plot_components(n, data, x_lpf, x_L1, u_L1, v_L1, txt_L1); orient tall printme('L1_components')
figure(1) clf subplot(2, 1, 1) plot(n, data) title('Data') xlabel('Time (samples)') axis(ax) subplot(2, 1, 2) plot( n, data, 'color', GRAY); line( n, x_L1 ); title('Sparsity-assisted signal smoothing (SASS) with L1 norm') xlabel('Time (samples)') axis(ax) orient portrait printme('L1')
SASS (log penalty)
The log penalty function is non-convex
% rho : Controls non-convexity of penalty (0 <= rho <= 1) rho = 0.5; % initialize with L1 solution [x_log, cost_log, u_log, v_log, a] = sass(y, d, fc, K, lam, 'log', rho, Nit, u_L1); txt_log = sprintf('SASS (log, K = %d, \\lambda = %.2f, \\rho = %.2f, %s)', K, lam, rho, txt_filt); err = s - x_log; RMSE_log = sqrt(mean(err(K+1:end-K).^2))
0 incorrectly locked zeros detected.
RMSE_log =
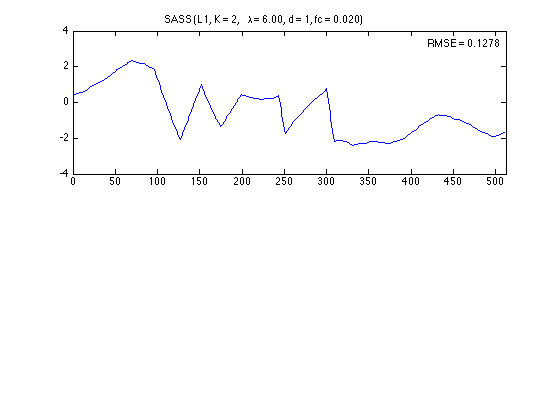
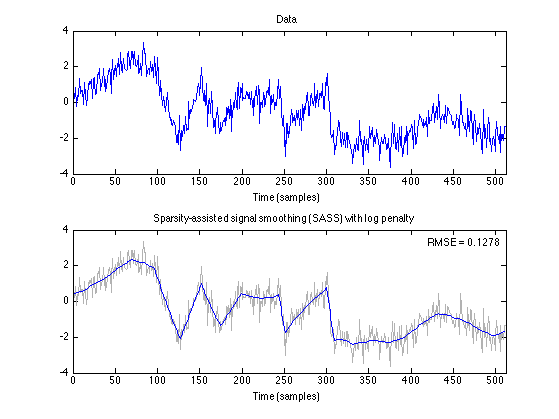
0.1278
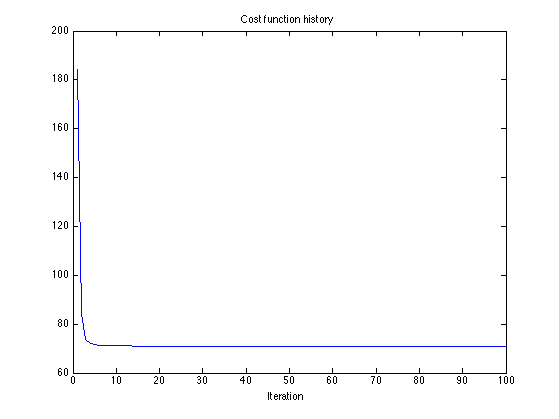
Cost function
figure(2) clf plot(cost_L1) title('Cost function history'); xlabel('Iteration')
Denoised signal
figure(2)
clf
subplot(2, 1, 1)
plot(n, x_log)
title(txt_L1)
axnote(sprintf('RMSE = %.4f', RMSE_log))
axis(ax)
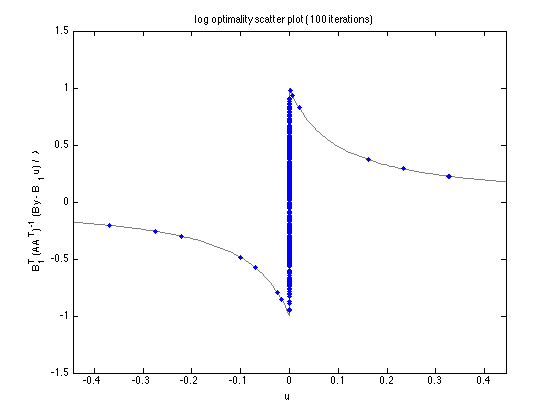
Optimality scatter plot
figure(2) clf plot_scatter(A, B, B1, y, u_log, lam, 'log', a); title(sprintf('log optimality scatter plot (%d iterations)', Nit)) printme('log_satter')
Components
figure(1) clf plot_components(n, data, x_lpf, x_log, u_log, v_log, txt_log); orient tall printme('log_components')
figure(1) clf subplot(2, 1, 1) plot(n, data) title('Data') xlabel('Time (samples)') axis(ax) subplot(2, 1, 2) plot( n, data, 'color', GRAY); line( n, x_log ); title('Sparsity-assisted signal smoothing (SASS) with log penalty') % title(txt_log) axnote(sprintf('RMSE = %.4f', RMSE_log)) xlabel('Time (samples)') axis(ax) orient portrait printme('log')
Save figure to pdf file
set(0, 'DefaultAxesFontSize', 8); figure(10) clf M = 4; subplot(M, 1, 1) plot(n, data) title('Noisy data'); axis(ax) subplot(M, 1, 2) plot(n, x_lpf); title(txt_LPF) axnote(sprintf('RMSE = %.3f', RMSE_LPF)) axis(ax) subplot(M, 1, 3) plot(n, x_L1) title(txt_L1); axnote(sprintf('RMSE = %.3f', RMSE_L1)) axis(ax) subplot(M, 1, 4) plot(n, x_log) title(txt_log); axnote(sprintf('RMSE = %.3f', RMSE_log)) axis(ax) orient tall printme('fig1') set(0, 'DefaultAxesFontSize', 'remove');